Removing duplicated records can be cumbersome. When collating bibliographic records from multiple literature databases both the total number of records and the proportion of duplicates can be high making manual removal of duplicates extremely time-consuming. Manual resolution of each set of potentially duplicated records is required when using reference managers such as Zotero or EndNote, and especially a screening platform Rayyan (note that deduplication algorithms available in all these are reasonably good at detecting (flagging) duplicating records (exact and non-exact duplicates), but not perfect, so combining different approaches is recommended anyway).
Here, I present an efficient workflow in which records from multiple sources (literature databases) are combined in Rayyan (https://rayyan.ai/), then automatically deduplicated using an R script (www.r-project.org), and finally uploaded into Rayyan again for the final round of deduplication and screening. Importantly, apart from Rayyan and R no other software is needed (but, at any stage, you can import/export lists of records into your reference manager to see the records or convert file formats). I assume you are already quite familiar with Rayyan and R.
The workflow:
1. Gather the bibliographic files.
Download lists of bibliographic references (with abstracts) from databases used to run the literature searches. Most of the time, exporting thema as a .ris file would work best. Rayyan has guidelines for the most commonly used databases on its upload page (see the screenshot below).
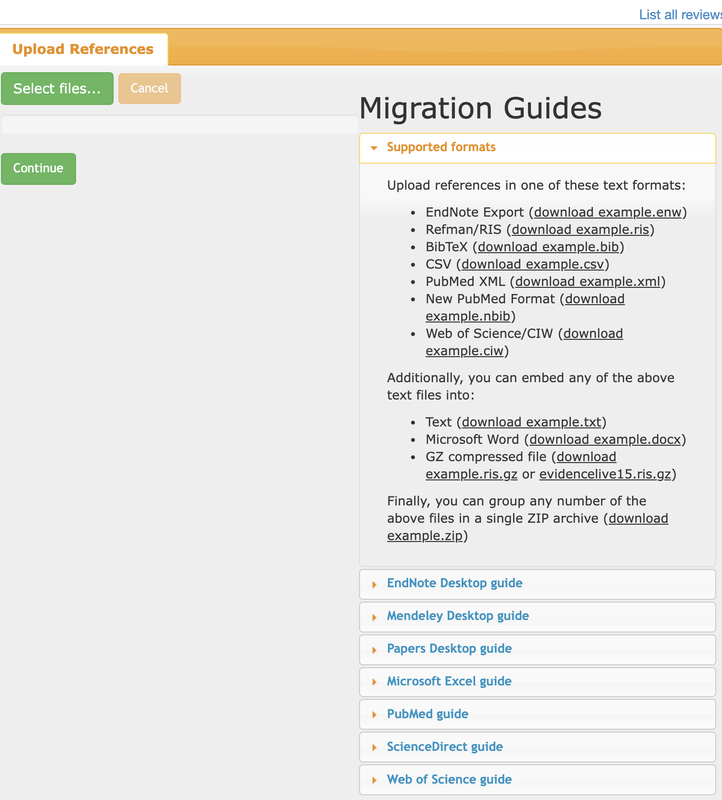
Create a new project in Rayyan and upload all files into it. This will create a combined list of records.
3. Run deduplication algorithm in Rayan (optional).
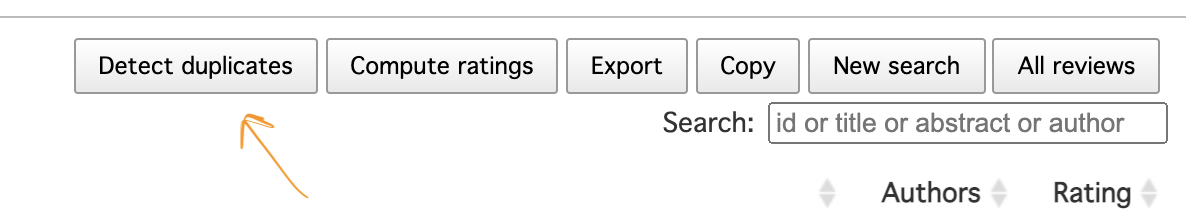
This will give you an idea on how many duplicated records you have in the combined set of records (if less <200 you may want to resolve them manually in Rayyan). To run the algorithm, press a “Detect duplicates” button close to the top right corner of the view with the list of combined references in Rayyan.
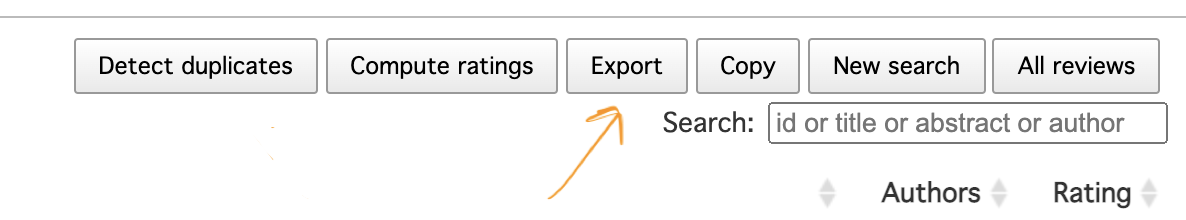
This will create one .csv with all references in the same format. To export the records, press a “Export” button close to the top right corner of the view with the list of combined references in Rayyan. In the pop-up window select “All” and “CSV” format (you can include all the fields listed below these options). Note that Rayyan will send you a link via email to download a compressed file. After decompressing, rename the .csv file to something usable (e.g., "FILENAME.csv") and place it in your R project folder.
Load the R packages needed:
library(tidyverse) # https://www.tidyverse.org/
library(synthesisr) # https://CRAN.R-project.org/package=synthesisr
library(revtools) # https://revtools.net/
dat <- read.csv("FILENAME.csv") #load the file
dim(dat) #see the initial number of uploaded references
6. Prepare data for deduplication in R.
We will deduplicate by comparing titles. Before doing so, it is good to tidy them up by bring them to the same case, removing extra white spaces and punctuation. We save these “processed” titles in a new column.
dat$title2 <- stringr::str_replace_all(dat$title,"[:punct:]","") %>% str_replace_all(.,"[ ]+", " ") %>% tolower() # Removing all punctuation and extra white spaces
7. Remove exact title matches in R.
This step uses processed titles to create a new smaller list of references with exact duplicates removed. It will save computational time for the next step (detection of non-exact duplicates).
dat2 <- distinct(dat, title2, .keep_all = TRUE) #reduce to records with unique titles
(removes exact duplicates)
dim(dat2) #see the new number of records
#View(arrange(dat2, title2)$title2) #an optional visual check - sorted titles
8. Deduplicate by fuzzy matching the remaining titles in R.
This step uses string distances to identify likely duplicates - it may take a while for long lists of references.
duplicates_string <- synthesisr::find_duplicates(dat2$title2, method = "string_osa", to_lower = TRUE, rm_punctuation = TRUE, threshold = 7)
#dim(manual_checks) #number of duplicated records found
#View( review_duplicates(dat2$title2, duplicates_string) # optional visual check of the list of duplicates detected. If needed, you can manually mark some records as unique (not duplicates) by providing their new record number from duplicates_string (duplicates have the same record number), e.g.
#new_duplicates <- synthesisr::override_duplicates(duplicates_string, 34)
dat3 <- extract_unique_references(dat2, duplicates_string) #extract unique references (i.e. remove fuzzy duplicates)
dim(dat3) #new number of unique records
9. Prepare the data for exporting from R.
Modify the data frame into a format that can be imported to Rayyan (the files saved as .bib or .ris for .csv files cannot be directly uploaded to Rayyan due to some formatting changes happening during processing them in R). This is done by first selecting only the key columns, saving them into a BibTex format (.bib file) and them changing the record labels into the desired format.
dat3 %>% select(key, title, authors, journal, issn, volume, issue, pages, day, month, year, publisher, pmc_id, pubmed_id, url, abstract, language) -> dat4 #select the key columns
write_refs(dat4, format = "bib", file = "FILENAME_deduplicated.bib") #save into a bib file
readLines("FILENAME_deduplicated.bib") %>%
stringr::str_replace(
pattern = "@ARTICLE",
replace = "@article") %>%
writeLines(con = " FILENAME_deduplicated.bib") #fix the record labels and save again as a .bib file
10. Import deduplicated records into Rayyan.
Create a new project in Rayyan and import the modified .bib file. Run the algorithm for detecting duplicates in Rayyan (see Point 3 above). This will reveal potential duplicates that were below the similarity threshold used in R (or have lots of formatting differences). These will need to be resolved manually in Rayyan (usually it is not a big number and some will require human intelligence to tell what counts as a real “duplicate”). After resolving these duplicates you are ready to start screening your deduplicated records in Rayyan.
Note: Unfortunately, record fields with authors and keyword information (and many other fields) are stripped from the original records in the above workflow, mostly by Rayyan. For this reason, records exported from Rayyan are usually not suitable for direct use in bibliometric analyses. But, at least, you can claim that your screening of bibliographic records in Rayyan was blinded to the authors’ identity.
 RSS Feed
RSS Feed