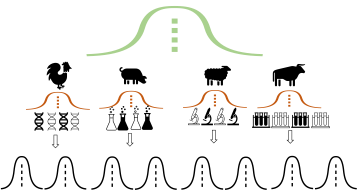
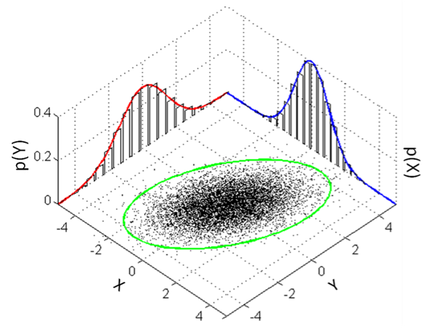
As I have been doing more surveys on meta-analytic practices in many disciplines and re-analysing more published meta-analysis (MA) papers, there is one “recommendation” that is growing stronger and stronger in my brain. That is, we should say goodbye to traditional fixed- and random-effects MAs and conduct our MAs using advanced methods like multilevel and multivariate models because meta-analytic datasets are often multilevel and multivariate in nature. Doing so can make sure you properly handle statistical issues like dependency, and heteroscedasticity, resulting in more robust parameter estimations and inferences. My main argument is that in the “worst-case” scenario, where your dataset does not have a complex structure thereof, these advanced models will automatically reduce into a normal fixed- and random-effects models, all with similar (or identical) results to those expected. More importantly, applying advanced methods can help you decompose variances (Figure 1) and separate correlations of true effects from observed effects (Figure 2), delivering new biological insights. I can see the between-study heterogeneity and correlation are overestimated in many published meta-analyses using fixed-and random-effects models.
|
By Yefeng Yang As I have been doing more surveys on meta-analytic practices in many disciplines and re-analysing more published meta-analysis (MA) papers, there is one “recommendation” that is growing stronger and stronger in my brain. That is, we should say goodbye to traditional fixed- and random-effects MAs and conduct our MAs using advanced methods like multilevel and multivariate models because meta-analytic datasets are often multilevel and multivariate in nature. Doing so can make sure you properly handle statistical issues like dependency, and heteroscedasticity, resulting in more robust parameter estimations and inferences. My main argument is that in the “worst-case” scenario, where your dataset does not have a complex structure thereof, these advanced models will automatically reduce into a normal fixed- and random-effects models, all with similar (or identical) results to those expected. More importantly, applying advanced methods can help you decompose variances (Figure 1) and separate correlations of true effects from observed effects (Figure 2), delivering new biological insights. I can see the between-study heterogeneity and correlation are overestimated in many published meta-analyses using fixed-and random-effects models. Although these advanced methods are good, there are (at least) three remarks worth noting here. First, all your models should be built strictly based on predefined questions (e.g., a priori hypotheses). Second, before applying these models, you need to correctly understand the statistical theory behind them. Otherwise, you very likely disseminate misleading information if you published results from them. Third (but not the last), do not use complex models to fit a small-sample-size dataset. This is especially true for multivariate models, which are often heavily parameterized (even overparameterized). So, always do (at least some basic) model checking (e.g., likelihood profile, convergence) to ensure stability of your model fitting. As I have been knowing more about statistics, I realised that many methods are just a special form of a more general framework. For example, (two-sample) Student t-test is a special form of ANOVA, which is a special form of linear regression, which is a special form of generalized linear model or linear mixed model, which is a special form of generalized linear mixed model, which is a special form of the generalized additive mixed model. In the same vein, fixed-effect MA is a special form of random-effects MA, which is a special form of a multilevel or multivariate model. I can imagine that one might disagree with “say goodbye to fixed- and random-effects meta-analyses”. For example, fixed-effects MA can still provide valid inferences if limiting your results to the included studies (e.g., conditional inference). I acknowledge this is true as long as you are not goanna generalize results beyond the included studies. I know asking people to resort to complex methods is difficult because people like easily-understandable tools - just think about P-value. I am always open and happy to see different ideas. Lastly, all the above claims only represent my personal intuition and opinion (I might extend it into a paper in future). They might be wrong and do not necessarily speak for my lab’s attitudes toward meta-analyses.
0 Comments
|
AuthorPosts are written by our group members and guests. Archives
June 2024
Categories |
 RSS Feed
RSS Feed